Molecular biology
Over the past two decades, several preclinical advancements have been made in the field of melanoma molecular biology [1-15]. The study of familiar cases has led to the identification of putative melanoma susceptibility genes (e.g. p16, p14ARF, see Familial Melanoma section). However, familiar melanomas represent approximately 5-10% of all melanoma cases, and sporadic melanomas rarely show the same molecular derangements found in familiar cases. Therefore, other molecular derangements must be involved in the pathogenesis of most melanomas.
Although sunlight is believed to play a key role in melanomagenesis (see Sun Exposure paragraph in the Risk Factors section), the cascade of molecular events leading to sporadic melanoma development is just beginning to be dissected. In particular, thousands of reports have been published describing molecular mechanisms underlying melanoma development, progression and resistance to conventional therapies. A few classical examples of molecular pathways whose alterations have been linked to melanoma malignant behavior are the following: tyrosine kinase receptors (TKR) pathways (e.g. VEGFR, HER, TGFBR), Ras / Raf / MEK / ERK pathway, PI3K / Akt / PTEN / mTOR pathway, cell cycle regulation pathways (Rb / p53 / p16INKA / p14ARF / HDM2), epigenetic gene expression regulation (DNA methylation, histone acetylation and microRNA), programmed cell death (apoptosis) pathways (e.g. death receptor pathway: FAS, TRAILR, TNFR; mitochondrial pathway: Bcl2 family), common apoptosis effectors (e.g. caspases), protein chaperoning and degradation (HSP, proteasome).
Recently, the implementation on a large scale of novel biotechnologies such as high-throughput platforms (e.g. gene microarray, proteomics) and genomic tools (e.g. RNA interference) has further increased the discovery pace in the field of molecular oncology, including melanoma research [7, 16-20]. The huge amount of data presently available has required the development of dedicated statistical and mathematical models to analyze and integrate the information generated. Moreover, the myriad of interactions between single molecules, regulatory circuits, different pathways, normal and malignant cells is making the understanding of cancer biology so complicated that a new discipline called "systems biology" has been dedicated to the integration of all this information [21].
In a translational medicine perspective [22], these advancements should lead to the identification of highly accurate diagnostic (i.e. related the presence or absence of minimal residual disease), prognostic (i.e. related to patient survival) and predictive (tumor response to therapy) biomarkers as well as to the identification of molecular targets suitable for the development of cancer specific drugs. Ultimately, this approach should lead to the holy grail of molecular oncology, which is the clinical implementation of the personalized treatment of cancer (see Molecularly targeted therapy section).
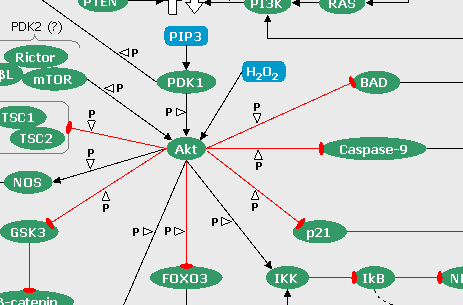
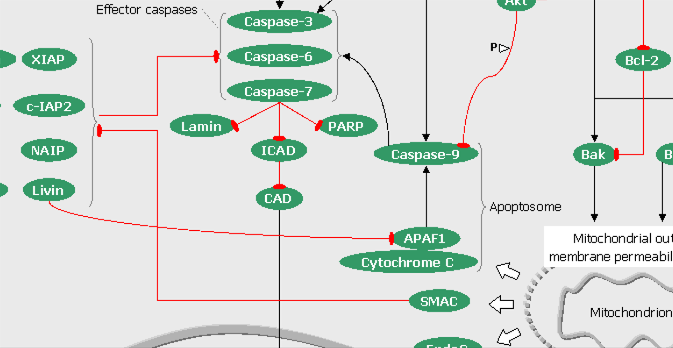
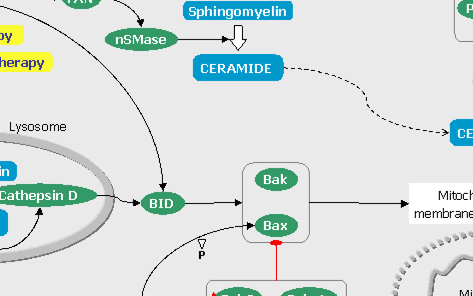
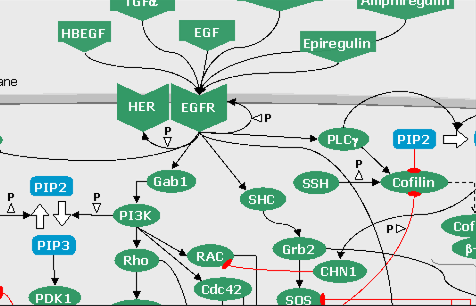
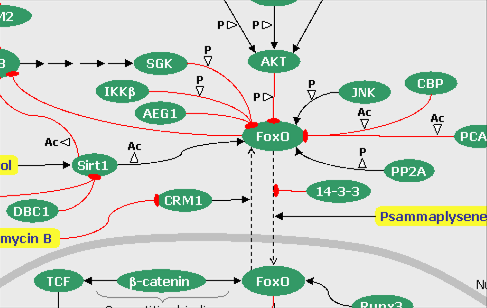
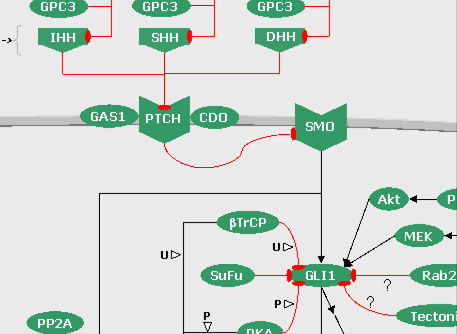
FIGURE 6: The Hedgehog pathways (click on the snapshot to open the corresponding Biomap).